This website uses cookies to ensure you get the best experience on our website.
- Table of Contents
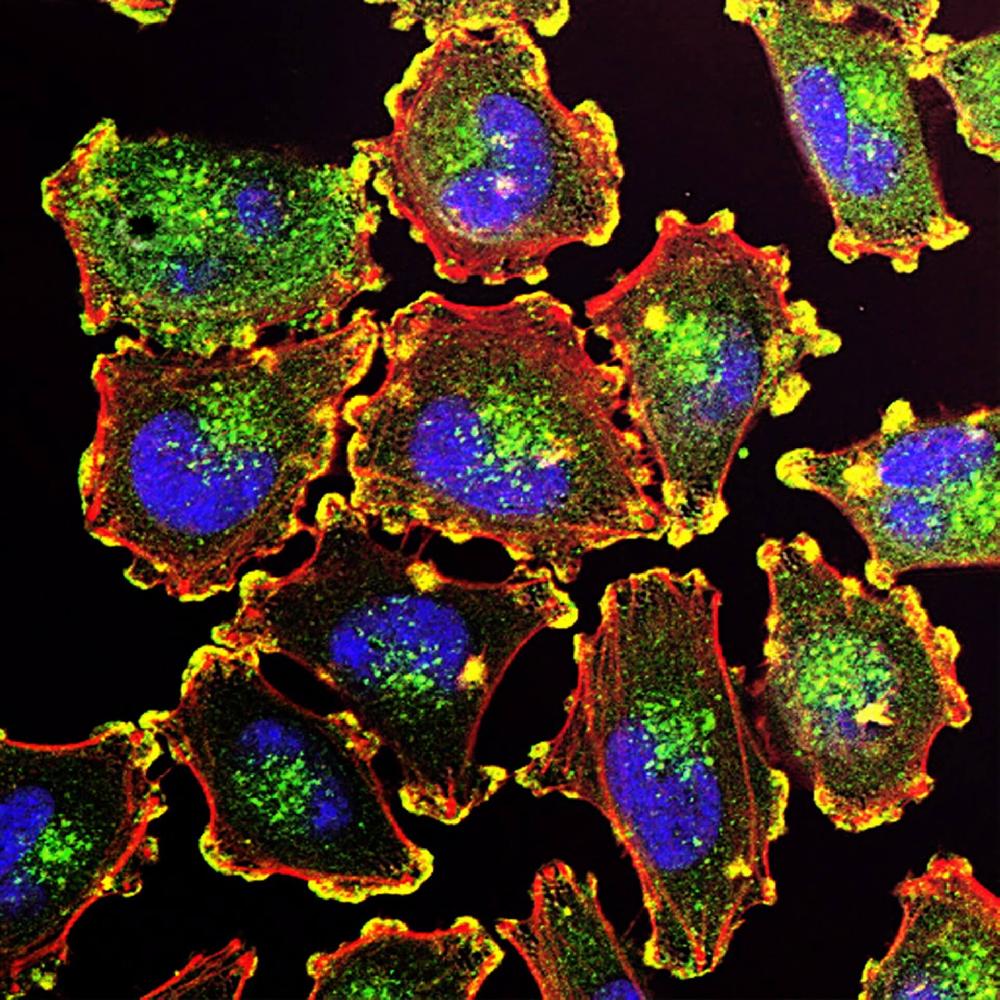
Facts about Terminal uridylyltransferase 7.

During 3' terminal uridylation of mRNA, sculpts, with TUT7, the maternal transcriptome by removing transcripts during oocyte growth (By similarity). Involved in microRNA (miRNA)- triggered gene silencing through uridylation of deadenylated miRNA targets (PubMed:25480299).
| Human | |
|---|---|
| Gene Name: | TUT7 |
| Uniprot: | Q5VYS8 |
| Entrez: | 79670 |

| Belongs to: |
|---|
| DNA polymerase type-B-like family |
Terminal uridylyltransferase 7
Mass (kDA):
171.229 kDA

| Human | |
|---|---|
| Location: | 9q21.33 |
| Sequence: | 9; NC_000009.12 (86287733..86354497, complement) |
Cytoplasm. Expression is pancytoplasmic in contrast with TUT4 expression which is enriched in cytoplasmic ribonucleoprotein granules.


PMID: 18172165 by Mullen T.E., et al. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5' to 3' and 3' to 5'.
PMID: 17353264 by Rissland O.S., et al. Efficient RNA polyuridylation by noncanonical poly(A) polymerases.